
In this example, we are not going to do that because we might want to use the headers later to label things, but keep this option in mind because you might want to use it in a later project. If we did this, then our values would all be numbers and we could use dtype=’float’, which is the default. The clever student may notice the skip_header option, where you can specify a number of lines to skip at the beginning of the file. In our file, our values are separated by commas we indicate that with delimiter=','. If you have mixed datatypes, like we do here, we want to use 'unicode'. For example, in our file, all the values were not numbers so we don’t want to use the datatype float, we want to use something else. We only need to specify these options if we don’t want to use the default value. All the other options have a default value that is shown after the = sign. We must put a values for this option because it does not have a default value. The first input fname is the filename we are reading in. The help menu shows us all the options we can use with this function. Load data from a text file, with missing values handled as specified.Įach line past the first `skip_header` lines is split at the `delimiter`Ĭharacter, and characters following the `comments` character are discarded. Genfromtxt(fname, dtype=, comments='#', delimiter=None, skip_header=0, skip_footer=0, converters=None, missing_values=None, filling_values=None, usecols=None, names=None, excludelist=None, deletechars=None, replace_space='_', autostrip=False, case_sensitive=True, defaultfmt='f%i', unpack=None, usemask=False, loose=True, invalid_raise=True, max_rows=None, encoding='bytes') We will use the help() function to learn more about genfromtxt() and how it works. One of these functions is the genfromtxt() function. The numpy library has several functions available to read in tabular data. In analyzing tabular data, we often need to perform the same types of calculations (averaging, calculating the minimum or maximum of the data set), so we are once again going to use a python library, this time a library that contains lots of functions to perform math operations. Open the file in a text editor and study it to determine its structure. This file was downloaded as part of your lesson materials. The table of atomic distances was saved as a CVS file called “distance_data_headers.csv”. These trajectories were generated with the AMBER molecular dynamics program and the distances were measured with the python program MDAnalysis. At each timestep, we are interested in the distance between particular atoms. The data was saved to the trajectory file every 1000 steps, so our file has 10,000 timesteps. We have a 20 ns simulation that used a 2 fs timestep. In this example, we have a CSV file that contains data from a molecular dynamics trajectory.
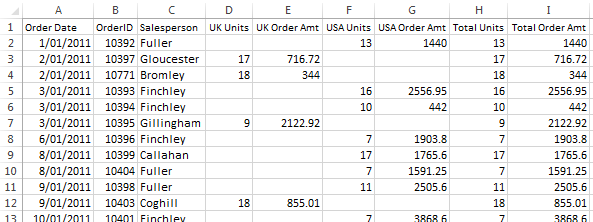
If you have data in a spreadsheet program that you need to import into a python code, you can save the data as a csvfile to read it in. Data is presented in rows, with each value separated by a comma. Frequently, a table will be mostly numbers, but have column or row labels.Ī common table format is the CSV file or comma separated values. But sometimes there are other ways that make more sense, particularly if the data is (1) all or mostly one type of data (for example, all numbers) and/or (2) formatted in a table. In theory, you could always use the readlines() function, and then use the data parsing tools we learned in the previous module to format the data as you needed.
In our last module, we used the readlines() function to read in a complex output file. Reading in Tabular DataĪs we already discussed, there are many ways to read in data from files in python. In this module we will focus on reading in and analyzing numerical data, visualizing the data, and working with arrays. Most scientists work with a lot of numerical data. Use 2D slices to work with particular rows or columns of data. Use functions in numpy to read in tabular data.


 0 kommentar(er)
0 kommentar(er)
